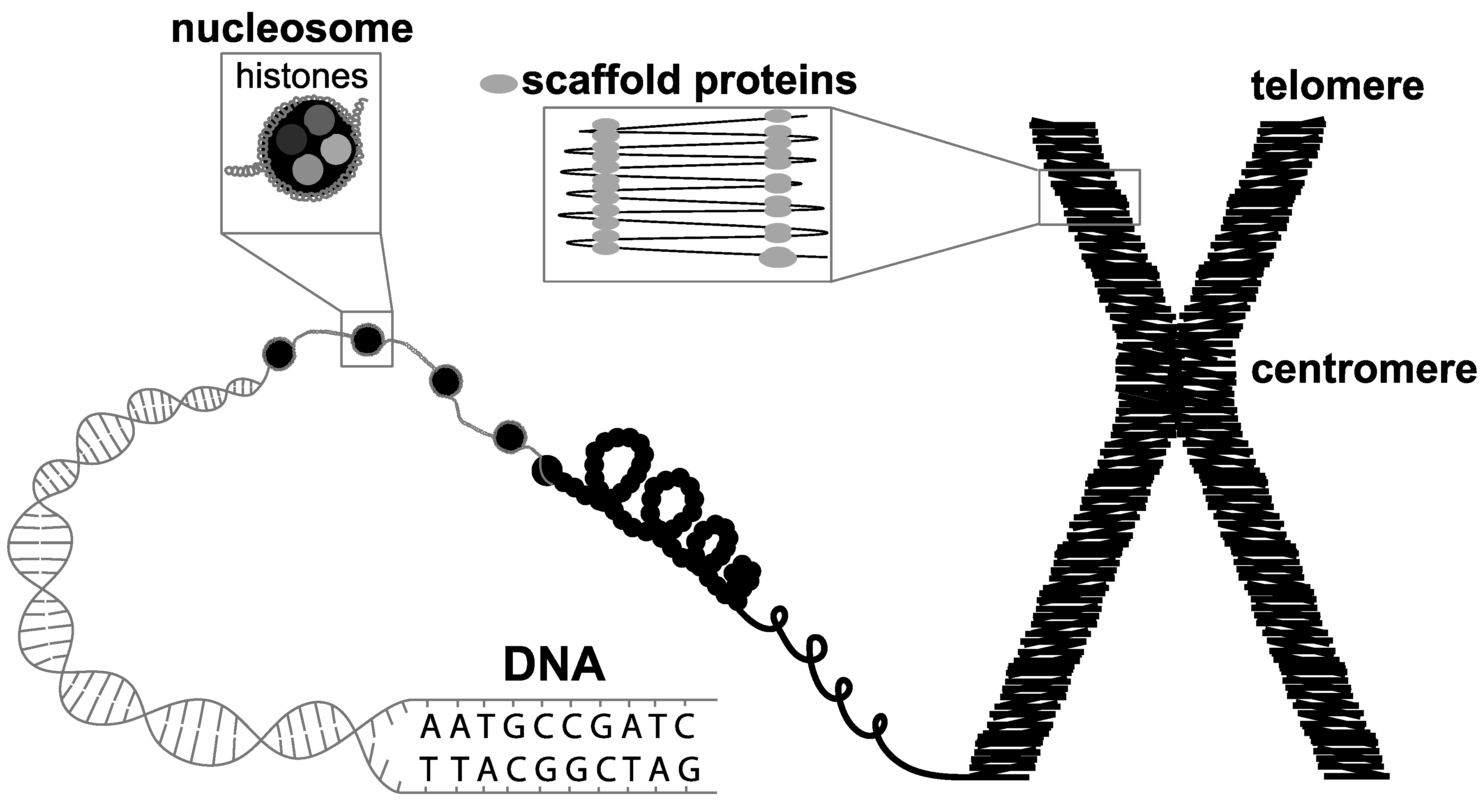
Detecting Biothreat Agents: From Current Diagnostics to Developing Sensor Technologies | ACS Sensors
Evaluation of molecular inversion probe versus TruSeq® custom methods for targeted next-generation sequencing | PLOS ONE
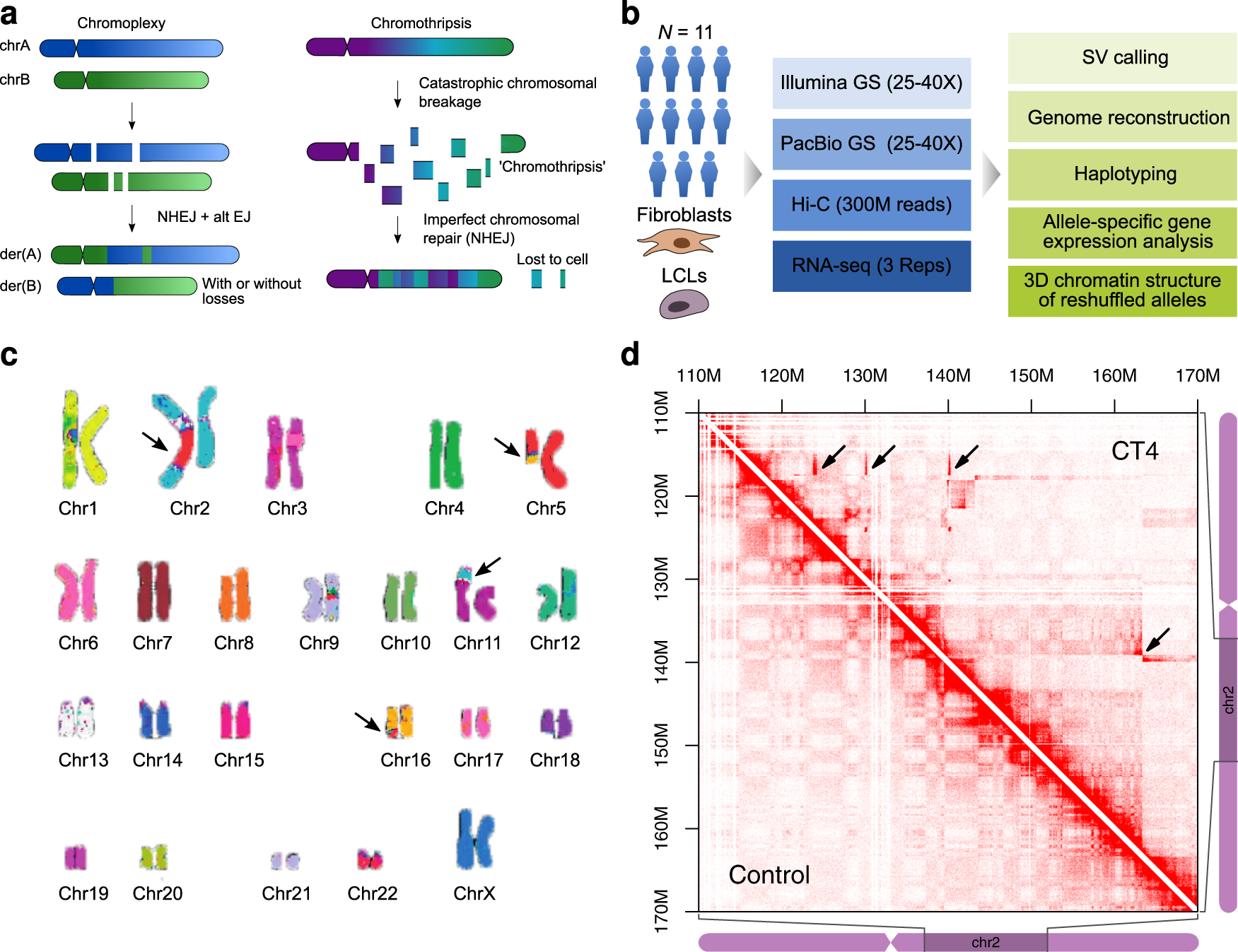
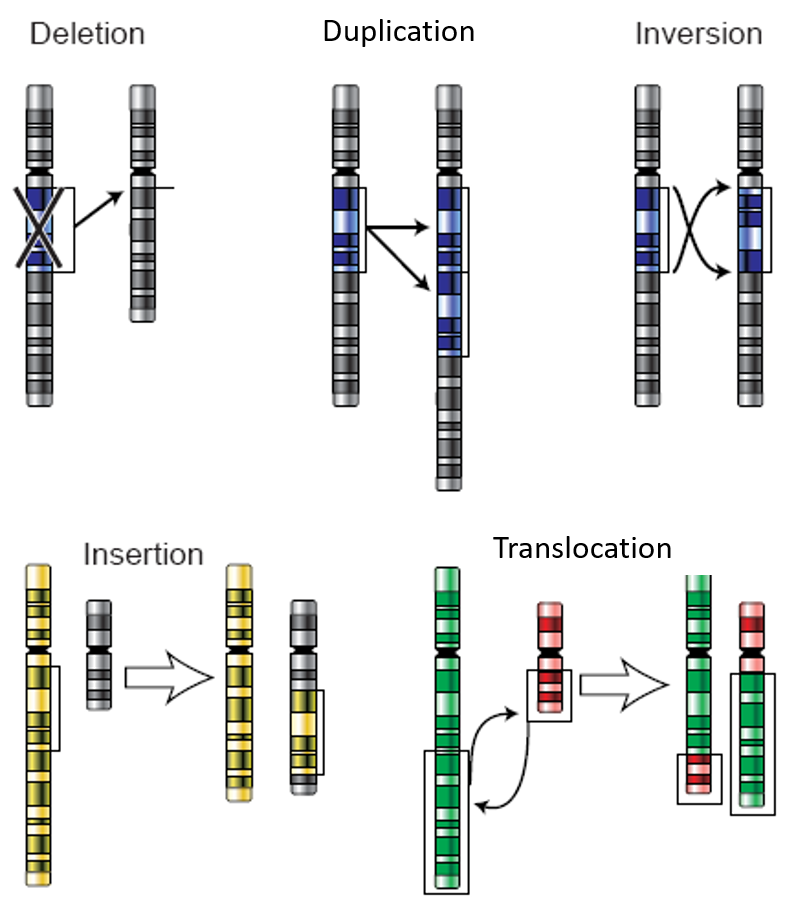
Integration of Hi-C with short and long-read genome sequencing reveals the structure of germline rearranged genomes | Nature Communications
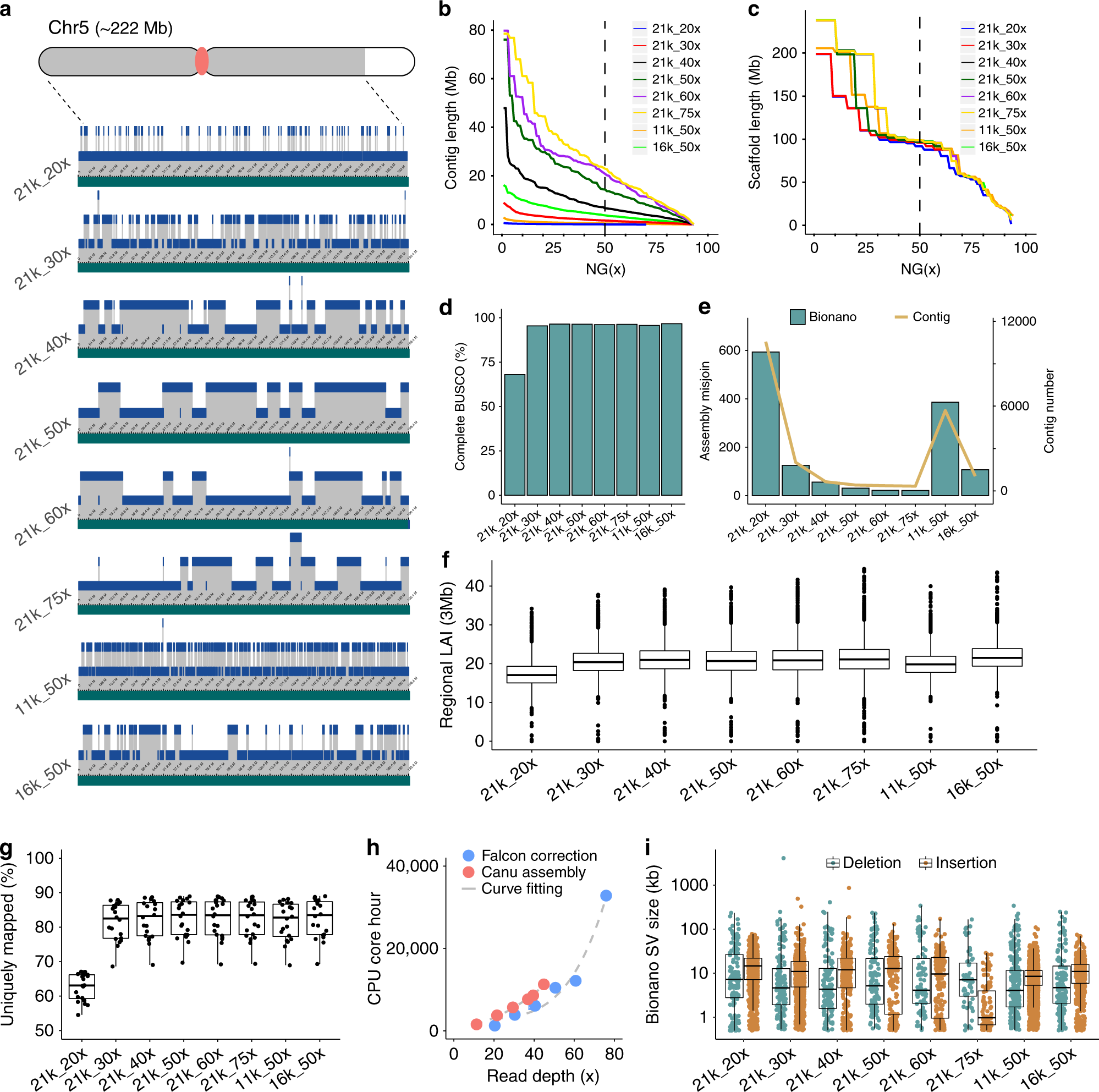
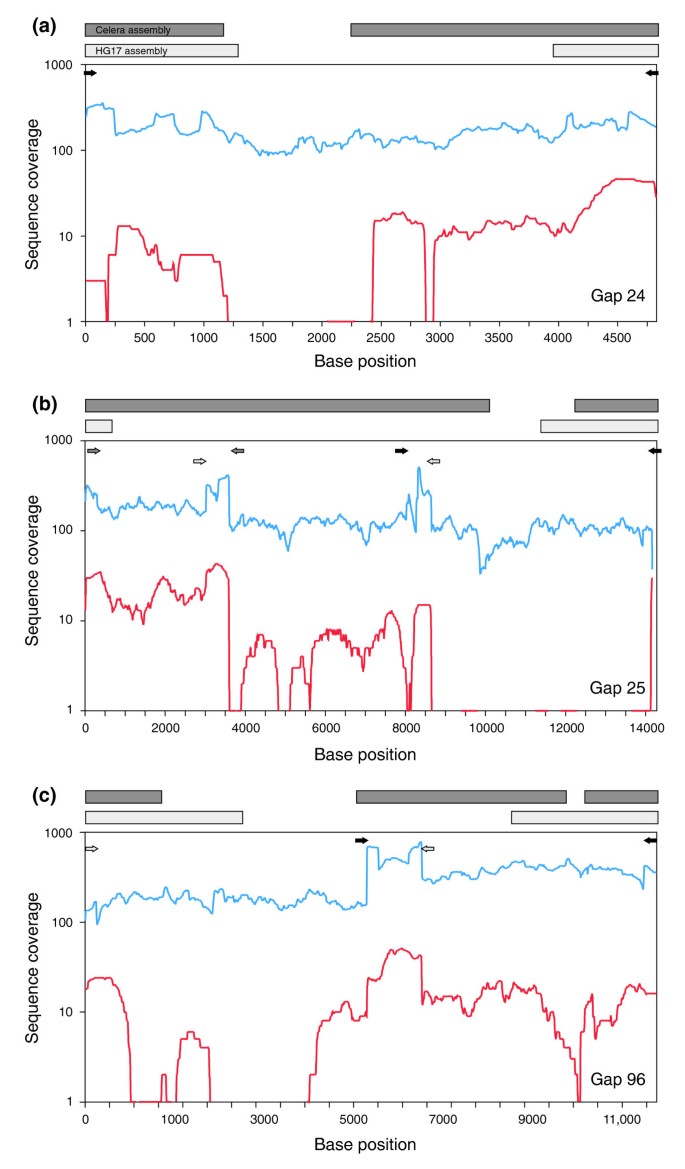
Effect of sequence depth and length in long-read assembly of the maize inbred NC358 | Nature Communications

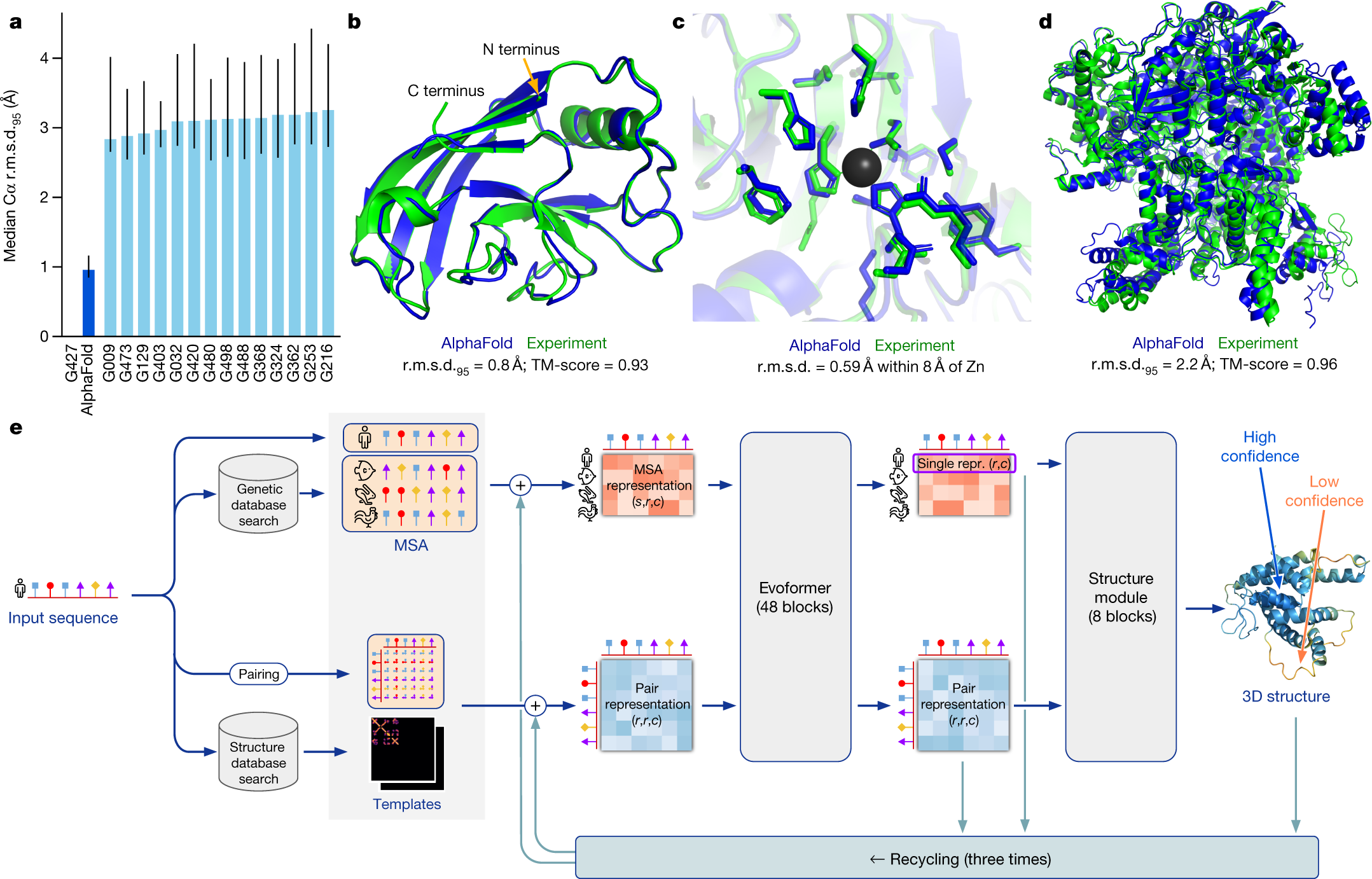
Molecular electronics sensors on a scalable semiconductor chip: A platform for single-molecule measurement of binding kinetics and enzyme activity | PNAS
Single strand gap repair: The presynaptic phase plays a pivotal role in modulating lesion tolerance pathways | PLOS Genetics

Figure 1 from Introducing Variable Gap Penalties into Three-Sequence Alignment for Protein Sequences | Semantic Scholar
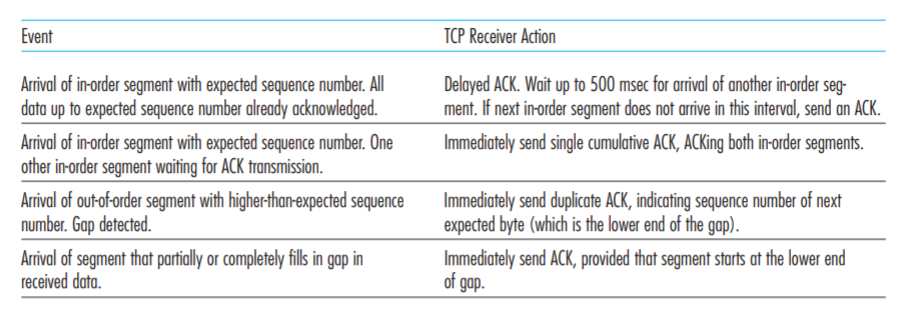
Calculate the Number of Gap Positions in a Sequence Alignment (and Coverage) · Issue #491 · biojava/biojava · GitHub